Introduction to functions and R packages
Mikhail Dozmorov
Virginia Commonwealth University
09-18-2023
Package priorities
Question: What is more important?
- Usability, solves real problem
- Statistical (methodological) superiority
- Documentation
- Speed
Documenting functions: the old way
- Originally, documentation was written in LaTeX-like format, stored in
man/*.Rdfiles
\name{cat_function}\alias{cat_function}\title{A Cat Function}\usage{cat_function(love.cats = TRUE)}\arguments{\item{love.cats}{Do you love cats? Defaults to TRUE.}}\description{This function allows you to express your love of cats.}\examples{cat_function()}\keyword{cats}Documenting functions: the simple way
- The package
roxygen2greatly simplifies documentation - Roxygen2 docstrings start with #’
Keywords defining pieces of documentation start with @
@param- parameter description@return- what the function returns@export- must be to make the function available@examples- how-to use the function
Can (must) use LaTeX syntax in special cases
\code{ <R code here> }- code highlight\url{ http:// ... }- URL\email{name@...}- e-mail
https://CRAN.R-project.org/package=roxygen2
Documenting functions: the simple way
- The package
roxygen2greatly simplifies documentation
#' A Cat Function#'#' This function allows you to express your love of cats.#' @param love.cats Do you love cats? Defaults to TRUE.#' @keywords cats#' @export#' @examples#' cat_function()Generating documentation
Run
roxygen2::roxygenise()ordevtools::document()to convert roxygen-formatted help to.Rdfiles understood by RCheck
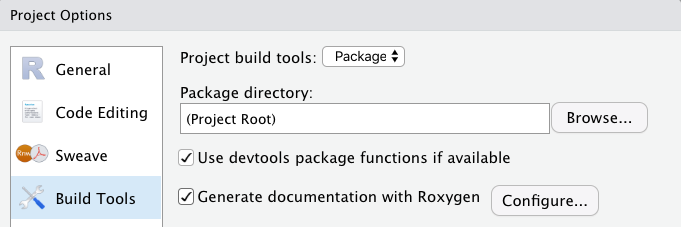
Generate documentation with Roxygento auto-generate.Rdfiles, NAMESPACE file. The menu "Tools -> Project Options -> Build Tools"
Making your functions available
- All packages have a
NAMESPACEfile: a collection of objects to be exported and imported- To avoid overwriting users' variables
- To avoid ambiguity in function calls
- To ensure the package has everything it needs to run
- To encourage modular code
# Generated by roxygen2: do not edit by handS3method(t,test2)export(TCGA_corr)export(Venn2)export(Venn3)export(Venn4)export(Venn5)export(gene_enrichment)Making your functions available
- A
NAMESPACEfile specifies which functions are available to the user, and which are hidden (helper functions, minimize naming conflicts)
export(function_name)- A minimal
NAMESPACEfile
# Export all namesexportPattern(".")- Your
NAMESPACEis auto generated using@export,@import,@importFromRoxygen tags; never directly modify yourNAMESPACEfile
Making objects from other packages available
- All or partial set of objects from another package can be imported and used as
package::object
import(randomForest)importFrom(ModelMetrics,mcc)importFrom(PRROC,pr.curve)- Your
NAMESPACEis auto generated using@export,@import,importFromRoxygen tags; never directly modify yourNAMESPACEfile
Making everything available with Roxygen2
Roxygen tags from function's help sections get converted to the NAMESPACE entries
In preciseTAD.R function:
#' @export#'#' @import randomForest e1071preciseTAD <- function(...)In NAMESPACE, after running roxygen2::roxygenise() or devtools::document()
export(preciseTAD)import(randomForest)import(e1071)Writing detailed documentation
Vignette – an instructive tutorial demonstrating practical uses of the software with discussion of the interpretation of the results (vignette = tutorial). Critical to get a user started with your package
A short introduction that explains
- The type of data the package can be used on
- The general purpose of the functions in the package
- One or more example analyses with
- A small, real data set
- An explanation of the key functions
- An application of these functions to the data
- A description of the output and how it can be used
https://github.com/hadley/dplyr/tree/master/vignettes
Writing vignettes
- Written using Markdown syntax
- Saved in
vignettes/*.Rmdfiles - Add YAML header to each vignette file
---title: "Vignette title"date: "2023-09-17"output: rmarkdown::html_vignettevignette: > %\VignetteIndexEntry{Vignette title} %\VignetteEngine{knitr::rmarkdown} %\VignetteEncoding{UTF-8}---- Build your vignettes with the
devtools::build_vignettes()command - The resulting
*.htmlfiles will be in theinst/docfolder
Package building pipeline using devtools
library(devtools)create(“cats”) # Create package skeletondocument(“cats”) # Create function's helpbuild_vignettes("cats") # Build vignettesbuild("cats") # Build packageinstall("cats") # Install packagecheck("cats") # Build and check a source package, using all known best practicesREADME.md for the package
- Create
README.Rmdwith the standard RMarkdown text and code, as you would do for the vignette - Use
devtools::build_readme()function that will compile theREADME.mdfile- Remember, the Markdown format in the
README.mdfile shouldn't have R code. - The
devtools::build_readme()function will execute code you put in theREADME.Rmdfile and format the code and its output properly
- Remember, the Markdown format in the
Package building pipeline using command line
R CMD build cats– will create a tarball of the package, with its version number encoded in the file nameR CMD install cats_0.0.0.9000.tar.gzR CMD check --as-cran cats_0.0.0.9000.tar.gz
Including datasets
Create
datafolderSave your data in R binary format, using
save(cats, file = “data/mydata.rda”)(or, use.RDataextension)Can include
.txtof.csvfilesAdd
LazyData: truein theDESCRIPTIONfile – your data will be immediately available (afterdata("mydata"),catsdata will be available on the first use).If the data is large, also add
LazyDataCompression: xz
Documenting datasets
- Add
R/mydata-data.Rfile - Document with
roxygen2syntax
#' My data brief info#'#' Longer description of my data#'#' @docType data#' @usage data(mydata)#' @format An object of class \code{"data.frame"}#' @keywords datasets#' @references Put reference here#' @source \href{http://....org}{Link}#' @examples#' data(mydata)"mydata" # No extensionExample of a dataset package
USDA Nutrients - an R package containing all data from the USDA National Nutrient Database, "Composition of Foods Raw, Processed, Prepared"
Use
devtools::install_github("hadley/usdanutrients")function to install a package from GitHub
https://github.com/hadley/usdanutrients
Updating R and packages
installr::updateR()- update R and the corresponding packages on WindowsupdateR- update R on Mac
Other useful tips and tricks
testthat is a H.W. package to write unit tests
rm(list=ls(all=TRUE))removes everything in the global environment- But does not unload packages! Use, e.g.,
detach("package:vegan", unload=TRUE) - Use "Session -> Restart R" to completely refresh your environment
- But does not unload packages! Use, e.g.,
pkgdown is a H.W. package that can autogenerate a website for your package
build_site()blogdown - Creating Websites with R Markdown, Yihui Xie et al.
bookdown - Write HTML, PDF, ePub, and Kindle books with R Markdown, by Yihui Xie et al.
More references
Writing R packages lecture slides by Daniel Sjoberg. GitHub source
How to create an R package lecture slides by Irene Steves, Mitchell Maier. GitHub source
R packages book by Hadley Wickham, GitHub